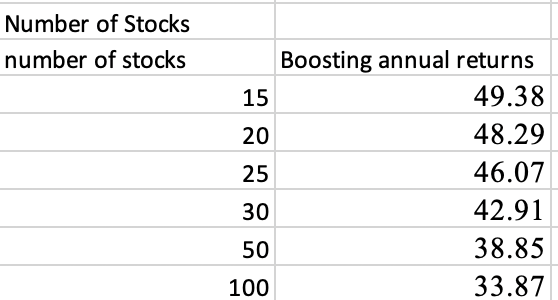
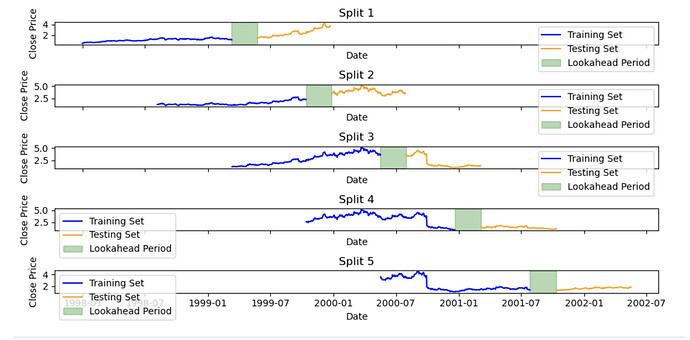
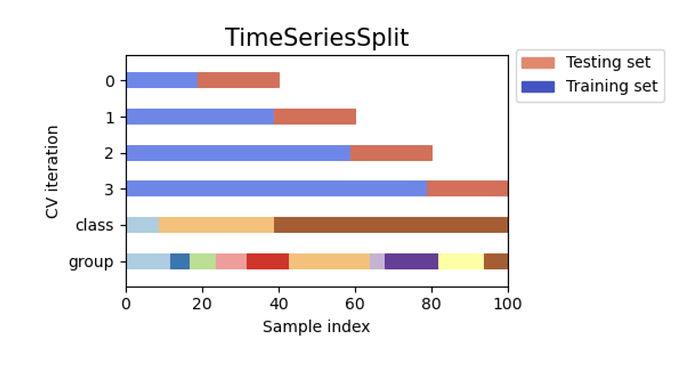
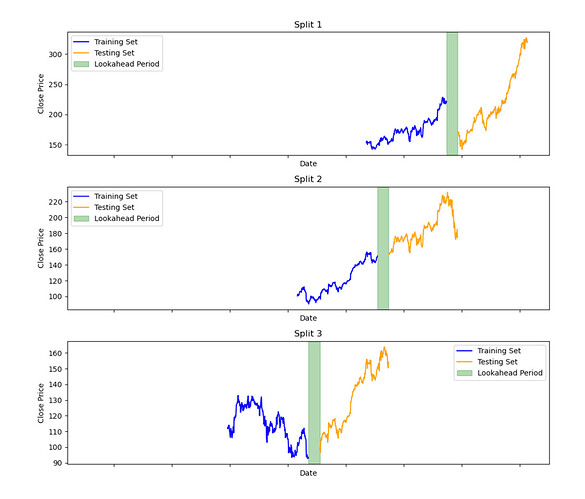
Not sure if it fits into this thread, but the name is ML workflow:
For my workflow I tested some data preprocessing on features.
The first was StandardScaler from scikit-learn, they say:
Quote
Standardization of datasets is a common requirement for many machine learning estimators implemented in scikit-learn; they might behave badly if the individual features do not more or less look like standard normally distributed data: Gaussian with zero mean and unit variance .
End quote
I did not found any effect.
Do I misunderstand something, how is you experience?
Here is the code:
scaler = StandardScaler()
# Fit the scaler on the training data and transform both training and test data
# Note: To maintain the multi-index and columns, we operate on the DataFrame values then reconstruct the DataFrame
# Scale X_train
X_train_scaled_values = scaler.fit_transform(X_train.values)
X_train = pd.DataFrame(X_train_scaled_values, index=X_train.index, columns=X_train.columns)
# Scale X_test using the same scaler
X_test_scaled_values = scaler.transform(X_test.values)
X_test = pd.DataFrame(X_test_scaled_values, index=X_test.index, columns=X_test.columns)
Second attempt should denoise the features, I found it in the book: de Prado, Marcos López . Machine Learning for Asset Managers, Chapter 2.5 Denoising.
Quote
It is common in financial applications to shrink a numerically ill-conditioned covariance matrix (Ledoit and Wolf 2004). By making the covariance matrix closer to a diagonal, shrinkage reduces its condition number. However, shrinkage accomplishes that without discriminating between noise and signal. As a result, shrinkage can further eliminate an already weak signal.
End quote
Maybe I programmed that wrong, it has as well no effect.
If someone has some experience, feedback would be greatly appreciated.
Here is the code:
def cov2corr(cov):
std = np.sqrt(np.diag(cov))
corr = cov / np.outer(std, std)
corr[corr < -1], corr[corr > 1] = -1, 1 # Numerical stability
return corr
def getPCA(corr):
pca = PCA()
pca.fit(corr)
return pca.explained_variance_, pca.components_.T
def denoisedCorr(eVal, eVec, nFacts):
# Copy and adjust the eigenvalues
eVal_ = np.diag(eVal).copy()
eVal_[nFacts:] = eVal_[nFacts:].sum() / float(eVal_.shape[0] - nFacts)
# Do not call np.diag on eVal_ again, as it is already a diagonal matrix
# Compute the denoised correlation matrix
corr1 = np.dot(eVec, eVal_).dot(eVec.T)
corr1 = cov2corr(corr1)
return corr1
def denoise(df,d):
# Handling NaNs and infinite values
df.replace([np.inf, -np.inf], np.nan, inplace=True)
df.fillna(0, inplace=True)
if d > 1:
d=1
nFacts = int(round(len(df.columns)*d,0))
#print('nFacts',nFacts)
# Assume df is your multi-index DataFrame with factors
corr_matrix = df.corr()
eVal, eVec = np.linalg.eigh(corr_matrix)
#nFacts = 10 # Example value, adjust based on your analysis
# Denoise the correlation matrix
denoised_corr = denoisedCorr(eVal, eVec, nFacts)
# Replace infinite values with NaNs
denoised_corr[np.isinf(denoised_corr)] = np.nan
# Fill NaNs with zero
denoised_corr = np.nan_to_num(denoised_corr)
do_plot = False
if do_plot:
denoised_eVal, _ = getPCA(denoised_corr)
# Ensure eigenvalues are sorted in descending order
original_eVal_sorted = np.sort(eVal)[::-1]
denoised_eVal_sorted = np.sort(denoised_eVal)[::-1]
# Plot the eigenvalues
plt.figure(figsize=(10, 7))
plt.semilogy(original_eVal_sorted, label='Original eigen-function', linestyle='-', linewidth=2)
plt.semilogy(denoised_eVal_sorted, label='Denoised eigen-function', linestyle='--', linewidth=2)
plt.ylabel('Eigenvalue (log-scale)')
plt.xlabel('Eigenvalue number')
plt.title('Comparison of Eigenvalues: Original vs Denoised')
plt.legend()
plt.show()
# Initialize PCA with the number of components you found relevant
pca = PCA(n_components=nFacts) # nFacts is the number of factors you decided to keep
pca.fit(denoised_corr)
# Transform the original factors
transformed_factors = pca.transform(df)
# Perform inverse transform to get denoised factors
denoised_factors = pca.inverse_transform(transformed_factors)
# Convert denoised factors back to DataFrame
denoised_df = pd.DataFrame(denoised_factors, index=df.index, columns=df.columns)
return denoised_df
.
I did as well some tests on the label, again not successful,
now going to enjoy 

Stuff is difficult
Please see:
Coqueret, Guillaume; Guida, Tony. Machine Learning for Factor Investing: Python Version (Chapman and Hall/CRC Financial Mathematics Series) (English Edition) (S.i). CRC Press. Kindle-Version.
Chapter 4
http://www.mlfactor.com/chap_4.html
from statsmodels.distributions.empirical_distribution import ECDF # to use the ECDF built in function
def norm_0_1(x):
return (x-np.min(x))/(np.max(x)-np.min(x))
def norm_unif(x):
return (ECDF(x)(x))
def norm_standard(x):
return (x- np.mean(x))/np.std(x)
.
Results:
norm_0_1 → no difference
norm_unif(y_train) → worse
norm_standard(y_train) → no difference
Im using a ExtraTreesRegressor.
If someone has some experience with Data preprocessing, feedback would be greatly appreciated - Thankx